Antibody sequence analysis is an essential step in the antibody discovery process. The bioinformatics tools that perform these analyses must often either be licensed or built separately from an existing informatics system.
Analysis of antibody sequences against germline or custom databases is built into Affinity's antibody discovery workflow, and Affinity's sequence analysis pipelines can be extended with your team's scripts. Spend time moving your discovery campaigns forward instead of getting multiple systems to cooperate in order to corral the data you need to make your decisions. Additional tools for searching Affinity's sequence database complement its analysis tools by rapidly honing in on sequences of interest and connecting them with information on associated proteins and plasmids.
Out of the box, Affinity analysis pipelines can execute NCBI's IgBlast against a reference database of choice or perform a germline comparison against your custom database. Variable chains identified through these analyses build up an internal database of variable regions and can be used to identify clonal populations from phage, hybridoma, and single B-cell campaigns. The variable regions database can also be populated with the output of other bioinformatics software, providing the flexibility to gather analyses from multiple sources. Both previously-performed analyses and the methods of Kabat and Chothia can be used to highlight antibody sequence elements, and scientists can record their own annotations on any sequence of interest to further elucidate key sequence elements for their team members.
Affinity's built-in sequence search and analysis tools provide a simple and intuitive means of working with sequence data without leaving the LIMS. Discovery teams can execute their campaigns from start to finish without the need for external tools, facilitating adoption by allowing those teams to hit the ground running.
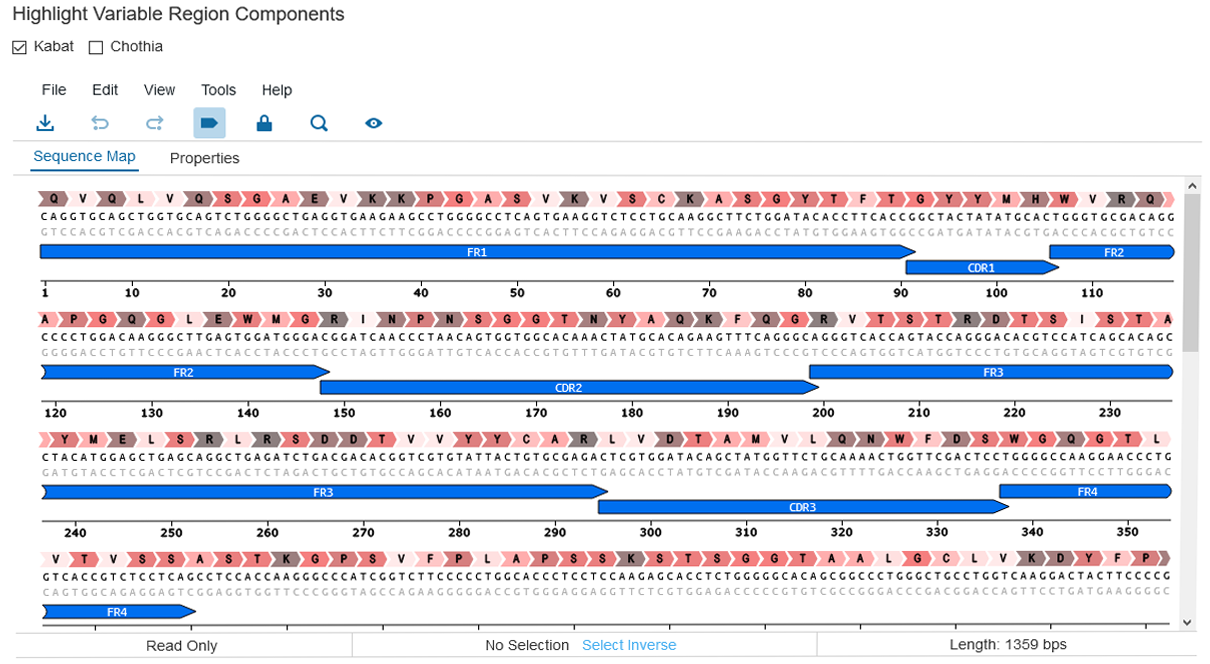
Intelligent sequence highlighting, use commercial or custom databases for reference
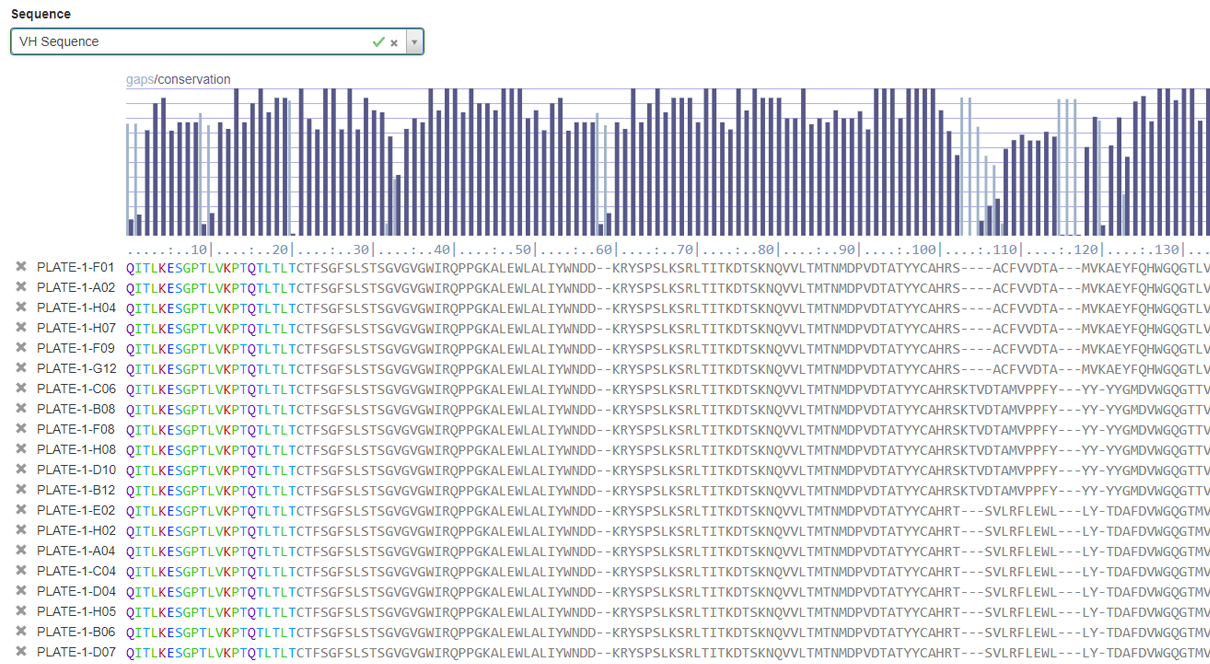
Powerful sequence alignment for specific regions of interest
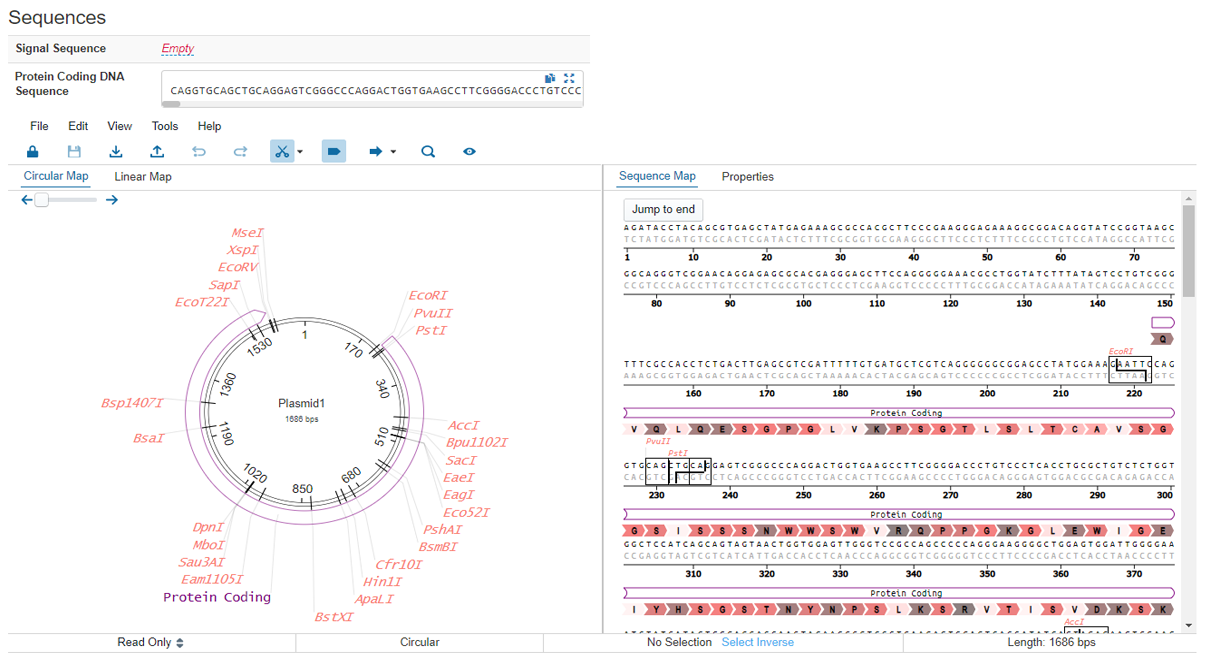
Modern plasmid sequence visualization tools


